Sigma-1 Receptor Modulation by Ligands Coordinates Cancer Cell Energy Metabolism
By Furkan E. Oflaz, Zhanat Koshenov, Martin Hirtl, Rene Rost, Roland Malli, and Wolfgang F. Graier
Excerpt from the article published in Biomolecules. 2022; 12(6):762. DOI https://doi.org/10.3390/biom12060762
Editor’s Highlights
- Sigma-1 Receptor (S1R) is an integral membrane protein, a unique ligand-activated chaperone, localized to the endoplasmic reticulum (ER) and highly enriched in specialized regions named mitochondria-associated ER membranes (MAMs).
- S1R is abundant in the Central Nervous System (CNS), but it is also highly expressed in liver, lung and different cancer cells [2].
- Under basal state, S1R was shown to interact with ER-resident chaperone BiP. Upon ER stress or agonist stimulation, S1R dissociates from BiP and interacts with many of its target proteins, including Inositol 1,4,5-trisphosphate receptors (IP3R).
- Cancer cells reprogram their energy metabolism to fit their demand for uncontrolled proliferation and survival.
- S1R is indispensable for enhancing mitochondrial bioenergetics during early ER stress by orchestrating ER Ca2+ leak towards mitochondria.
- Ligand-activated S1R increases bradykinin-induced cytosolic Ca2+ rise in neuroblastoma cells by dissociating adaptor protein Ankyrin from IP3Rs.
- Activation of S1R via agonist stimulation leads to the interaction of S1R with IP3Rs and controls the prolonged Ca2+ delivery to the mitochondria.
- S1R antagonist stabilizes IP3Rs and decreases cytosolic pyruvate/lactate ratio.
Abstract
Sigma-1 receptor (S1R) is an important endoplasmic reticulum chaperone with various functions in health and disease. The purpose of the current work was to elucidate the involvement of S1R in cancer energy metabolism under its basal, activated, and inactivated states. For this, two cancer cell lines that differentially express S1R were treated with S1R agonist, (+)-SKF10047, and antagonist, BD1047. The effects of the agonist and antagonist on cancer energy metabolism were studied using single-cell fluorescence microscopy analysis of real-time ion and metabolite fluxes. Our experiments revealed that S1R activation by agonist increases mitochondrial bioenergetics of cancer cells while decreasing their reliance on aerobic glycolysis. S1R antagonist did not have a major impact on mitochondrial bioenergetics of tested cell lines but increased aerobic glycolysis of S1R expressing cancer cell line. Our findings suggest that S1R plays an important role in cancer energy metabolism and that S1R ligands can serve as tools to modulate it.
1. Introduction
Sigma-1 Receptor (S1R) is an integral membrane protein localized to the endoplasmic reticulum (ER) and highly enriched in specialized regions named mitochondria-associated ER membranes (MAMs) [1]. S1R is abundant in the Central Nervous System (CNS), but it is also highly expressed in liver, lung and different cancer cells [2]. Under basal state, S1R was shown to interact with ER-resident chaperone BiP [1,3]. Upon ER stress or agonist stimulation, S1R dissociates from BiP and interacts with many of its target proteins, including Inositol 1,4,5-trisphosphate receptors (IP3R) [1]. Through the latter interaction, S1R can regulate efficient Ca2+ delivery to mitochondria. Moreover, agonist activation of S1R can lead to translocation of S1R to the cell membrane, where it regulates the activity of ion channels [4]. Consequently, S1R plays an important role in ion channel activity [4], apoptosis [5], and proliferation [6].
Localizing in different integral membranes, interacting with diverse classes of proteins, and activation-dependent regulation of different cellular pathways make S1R a potential target for metabolic modulation of cancer cells. S1R is highly expressed in several cancers including prostate, colon, melanoma, breast, and lung [7,8]. Several studies investigated the effects of S1R ligands in different cancer cell lines [5,6,7,8,9]. Among lung, breast, and prostate cancer cell lines, the highest expression of S1R was found in lung then breast, and colon cancer cell lines [6]. In these studies, an agonist of S1R, SKF10047, reduced the proliferation of high S1R expressing MDA-MB-231 breast cancer cell line but not of MCF-7 or MCF-10A cell lines, which have low levels of S1R expression [6]. Moreover, the S1R antagonist, Rimcazole, promotes caspase-dependent apoptosis in cancer cells and this effect was attenuated by S1R agonist SKF10047 [5]. Altogether these studies suggest that S1R expression, ligand activation, and function can determine cancer cell fate.
In this study, we investigated the role of S1R under basal, activated, and inactivated states on cancer energy metabolism by live-cell imaging of metabolic fluxes and ions in mitochondria and cytosol. Our results reveal that pharmacological activation of S1R increases mitochondrial bioenergetics by increasing basal Ca2+ delivery to mitochondrial matrix in highly S1R expressing A549 cells.
…
3. Results
3.1. Sigma-1 Receptor Is Differentially Expressed in A549, Lung Carcinoma, and MCF7, Breast Cancer, Cell Lines
For this study we have chosen two human cancer cell lines that differentially express S1R protein [14]. A549 lung adenocarcinoma cell line expresses S1R, while MCF7 breast cancer cell line expresses barely detectable levels of S1R (Figure 1A,B), and thus serves as a good control cell line. Additionally, the knockdown (KD) of S1R in A549 cells using small interfering RNA (siRNA) resulted in a 25% reduction of S1R protein level when analyzed without transfection selection marker (Figure 1B). As we have recently shown, this knockdown efficiency does not reflect the extent of protein downregulation in positively transfected single cells [15]. Hence, we have quantified protein downregulation in transfection positive sorted A549 cells and saw a 55% downregulation of S1R protein (Figure 1C,D). Since overall transfection efficiency of both cell lines using our transfection protocol is around 30–40%, we have relied on single-cell experiments with transfection selection markers for experiments with S1R KD.
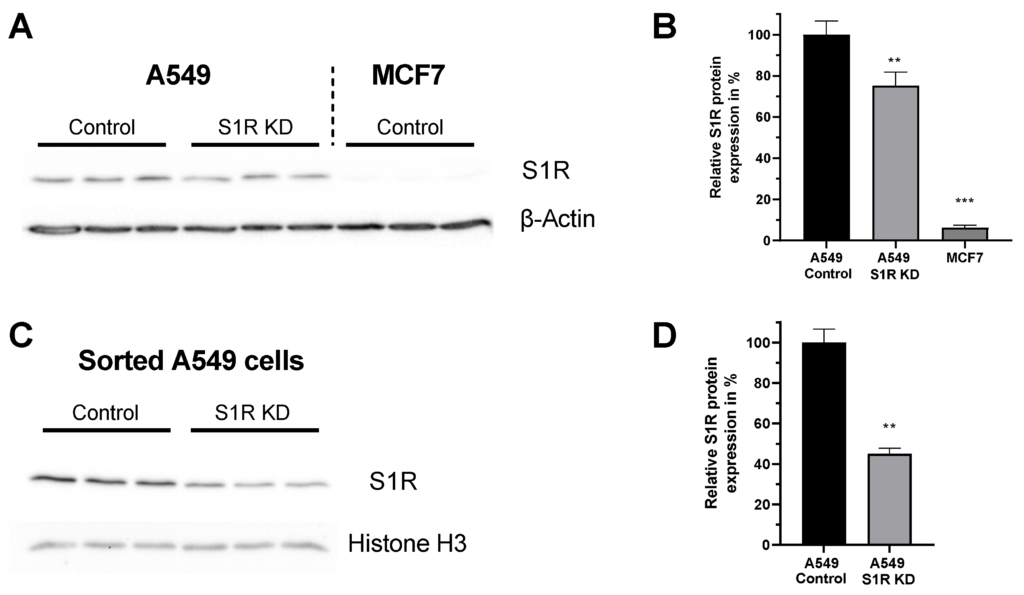
Expression level of S1R in A549 and MCF-7 Cells.
(A) Immunoblot images show the expression level of S1R in cells transfected either with control siRNA or siRNA against S1R (left) in A549 cells and non-transfected MCF7 cells (right). (B) Bar graphs represent immunoblot analysis of S1R expression as MEAN±SEM in A549 cells transfected with control siRNA or siRNA against S1R and non-transfected MCF7 cells. (C) Immunoblot images show the expression level of S1R in cells transfected either with control siRNA or siRNA against S1R in sorted, transfection positive A549 cells. (D) Bar graphs represent immunoblot analysis of S1R expression as MEAN±SEM in sorted A549 cells. Significant differences were assessed using one-way ANOVA with Tukey’s multiple comparison test (for B) or unpaired t-test (for D) and presented as (** p < 0.01, *** p < 0.001). A549 cells: A549 Control (n = 3), A549 S1R KD (n = 3), MCF7: (n = 3), sorted A549 Control (n = 3), sorted A549 S1R KD (n = 3).
3.2. Pharmacological Activation of S1R Increases Mitochondrial Bioenergetics While the Antagonist Doesn’t Affect It
To dissect the roles of S1R in cellular energy metabolism under basal, activated, and inactivated states we have performed real-time mitochondrial ATP measurements using genetically encoded mitochondrial ATP biosensor mtAT1.03 [10] (Figure 2A(i–iii)). We have deployed a protocol previously published by our group [16,17], which consists of two main steps: first, the glucose removal step, gives information on the reliance of the cells on glucose metabolism and mainly reflects glycolytic ATP flux, while the second step, oligomycin addition following re-introduction of glucose, reflects mitochondrial ATP production, thus oxidative phosphorylation (OXPHOS) level. The ratio of the second (oligomycin addition) to the first (glucose removal) step reflects the metabolic state of a cell and its reliance on OXPHOS versus glycolysis (Figure 2A(i–iii)). Activation of S1R using its well-described agonist, SKF-10047 [18], resulted in enhanced mitochondrial bioenergetics of A549 cells and increased their reliance on mitochondrial versus glycolytic ATP production (Figure 2B). The application of the specific S1R antagonist, BD-1047 [19], did not have a major effect on the metabolic state and mitochondrial bioenergetics of these cells (Figure 2B). The effect of (+)-SKF-10047 was abolished by the KD of S1R in A549 cells (Figure 2C) and neither S1R agonist nor antagonist had any effect in MCF7 cells (Figure 2D), suggesting S1R specificity of activation by (+)-SKF-10047. These findings point at OXPHOS promoting action of pharmacological S1R activation, while inactivation does not yield much of an effect, implying that S1R is mostly in its inactive form under the resting condition in regards to cellular energy metabolism.
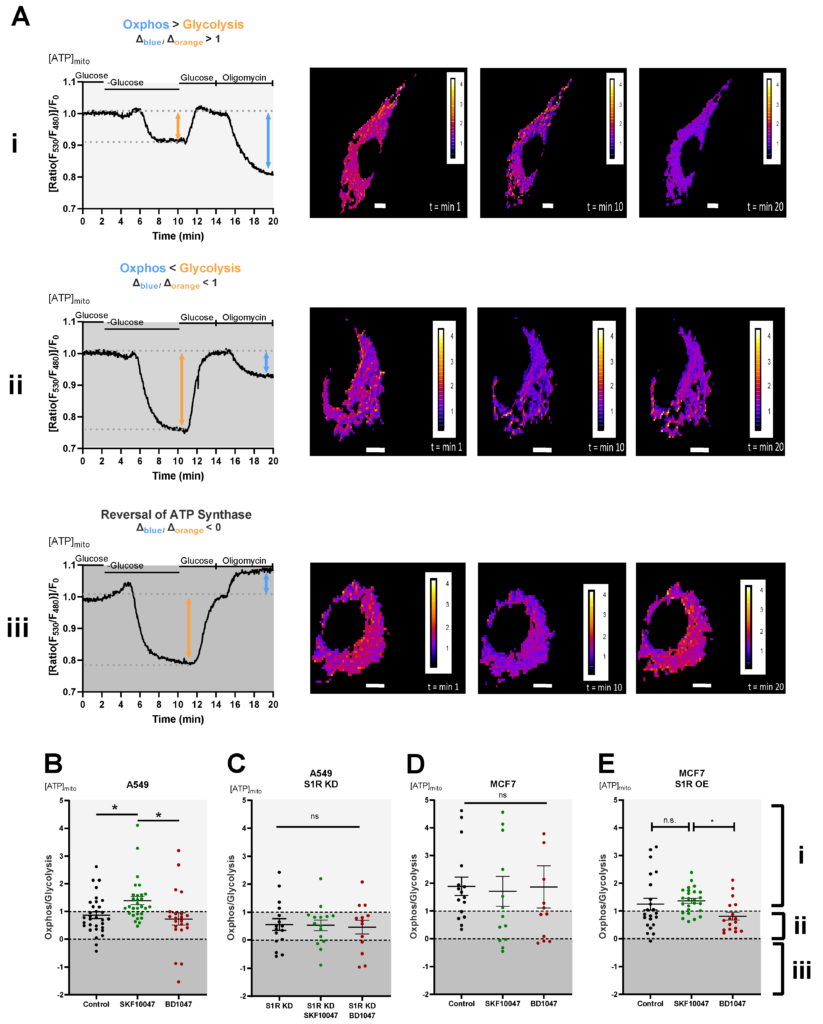
Effect of pharmacological activation of S1R on mitochondrial energy status.
(A) Representative traces of mitochondrial ATP levels with corresponding representative images of the cells transfected with mtAT.1.03 in MCF7 (i) and A549 (ii,iii) cells. The cells have been pseudocolored to represent mitochondrial ATP level as mtAT1.04 ratio; calibration bars are inserted on the right-hand side of the cells. The inserted white scale bar represents 10 µm. Change in ratio of oligomycin addition to glucose deprivation was used as a Oxphos/Glycolysis ratio and presented as Oxphos > Glycolysis (A(i)), Oxphos < Glycolysis (A(ii)), and reversal of ATP synthase (A(iii)). White area in panels (B–E) (indicated with (i)) represents Oxphos > Glycolysis, light gray area in panels (B–E) (indicates with (ii)) represents Oxphos < Glycolysis, and dark gray area in panels (B–E)(indicated by (iii)) represents reversal of ATP synthase. Scatter plots with individual values represent the ratio of Oxphos to Glycolysis in single cells with MEAN±SEM for (B) Control (black), Control+SKF10047 (green), Control+BD1047 (red), (C) S1R KD (black), S1R KD +SKF10047 (green) and S1R KD +BD1047 (red) in A549 cells, (D) for Control (black), Control+SKF10047 (green), Control+BD1047 (red) in MCF7 cells, and (E) for Control (black), Control+SKF10047 (green), Control+BD1047 (red) in MCF7 cells with S1R OE. Cells were treated with BD1047 and SKF10047 2–4 h prior to each experiment. Significant differences were assessed using one-way ANOVA with Tukey’s multiple comparison test and presented as (* p < 0.05, ns: not significant). A549 cells: Control (32 cells/16 experiments), Control + SKF10047 (29 cells/15 experiments), Control + BD1047 (22 cells/13 experiments), SR1 KD (16 cells/10 experiments), S1R KD + SKF10047 (15 cells/10 experiments) and S1RKD + BD1047 (13 cells/8 experiments). MCF7 cells: Control (15 cells/6 experiments), Control + SKF10047 (13 cells/4 experiments), Control + BD1047 (11 cells/5 experiments), S1R OE (22 cells/7 experiments), S1R OE + SKF10047 (25 cells/8 experiments), and S1R OE + BD1047 (18 cells/5 experiments).
To further validate these results, we have overexpressed (OE) S1R in MCF7 cells (Supplementary Figure S1), and performed the same protocol. Although the OE of S1R-mCherry construct altered overall metabolic response of the MCF7 cells, (+)-SKF-10047 had a clear OXPHOS promoting effect in comparison to BD1047 treated cells (Figure 2E).
In contrast to oligomycin addition glucose deprivation ratio readout, analysis of basal ATP level did not provide a significant difference upon agonist or antagonist treatment (Supplementary Figure S2).
3.3. S1R Agonist Increases Mitochondrial Membrane Potential and Reduces Aerobic Glycolysis, While the Antagonist Increases Aerobic Glycolysis
To further corroborate the results obtained with real-time mitochondrial ATP measurements without transfecting the cells, we deployed mitochondrial membrane potential (Ψm) assessment with a tetramethylrhodamine methyl ester (TMRM), a cationic fluorescent dye that accumulates in mitochondria depending on membrane potential (Figure 3A). In support of the previous experiment, activation of S1R with (+)-SKF10047 increased, while BD1047 did not affect Ψmin A549 cells (Figure 3B). Similar to ATP experiment, neither of the compounds changed Ψm in MCF7 cells (Figure 3C). As Ψm is a reflection of mitochondrial energetic status and directly represents cellular OXPHOS, S1R has an important role in promoting OXPHOS upon activation, while under basal state, it seems to be dormant in regards to this particular function, since there was no effect upon antagonist application.
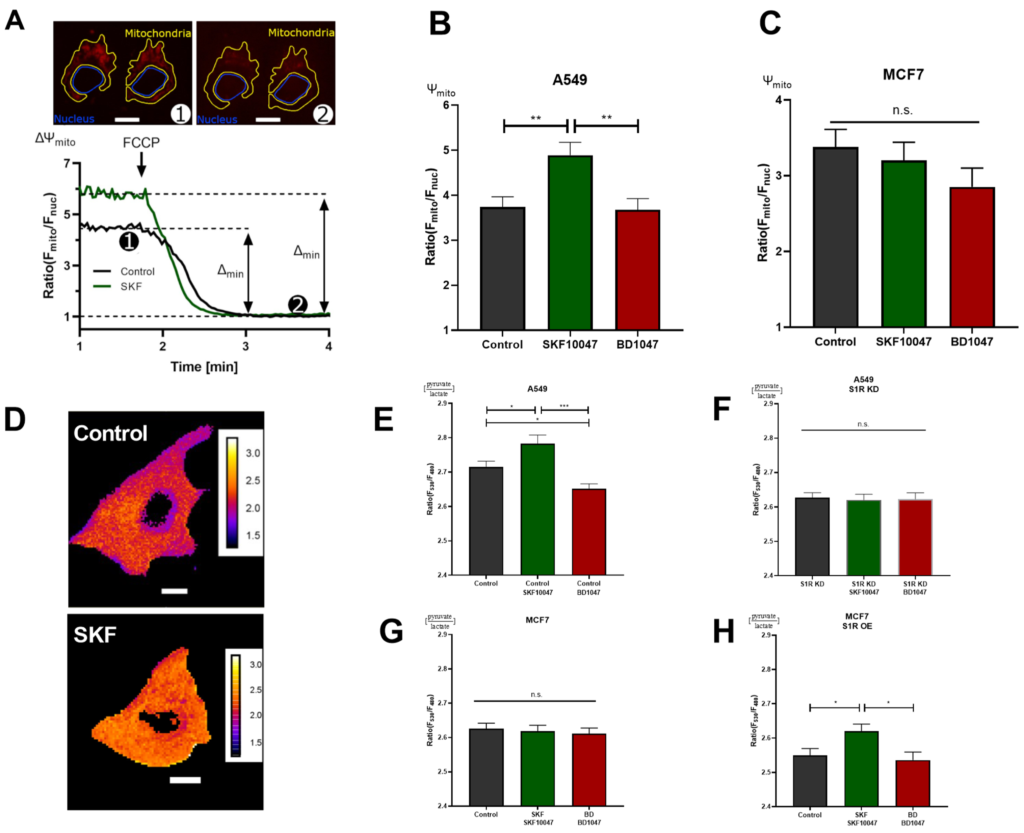
S1R ligands modulate cancer cell energy metabolism. (A) Exemplary images of A549 cells stained with TMRM and selected regions of interest (ROI) for mitochondria and nucleus. The inserted white scale bar represents 10 µm. The experimental protocol with representative ratio traces of mitochondrial to nucleus ROIs is shown for control (black) and control+SKF10047 (green) are shown. Protocol indicated in (A) was used to obtain mitochondrial membrane potential for Control (black) and Control+SKF10047 (green) by calculating the change in fluorescence ratio in mitochondria and in nucleus after 1 µM FCCP treatment. (B) Bar graphs with MEAN±SEM represent the mitochondrial membrane potential for Control (black), Control+SKF10047 (green) and Control+BD1047 (red) in A549 cells and (C) in MCF7 cells. (D) Exemplary images of A549 cells transfected with ratiometric pyruvate/lactate sensor for Control (above) and Control+SKF10047 (below). The cells have been pseudocolored to represent pyruvate/lactate ratio, calibration bars are inserted on the right-hand side of both cells. The inserted white scale bar represents 10 µm. (E) Bar graphs with MEAN±SEM represent the cytoplasmic ratio of pyruvate to lactate for Control (black), Control+SKF10047 (green), Control+BD1047 (red), (F) S1R KD (black), S1R KD +SKF10047 (green) and S1R KD +BD1047 (red) in A549 cells, (G) Control (black), Control+SKF10047 (green), Control+BD1047 (red) in MCF7 cells, and (H) Control (black), Control+SKF10047 (green), Control+BD1047 (red) in MCF7 cells with S1R OE. Cells were treated with BD1047 and SKF10047 2–4 h prior to each experiment. Significant differences were assessed using one-way ANOVA with Tukey’s multiple comparison test and presented as (* p < 0.05, ** p < 0.01, *** p < 0.001, ns: not significant). A549 cells TMRM measurements: Control (90 cells/4 experiments), Control + SKF10047 (114 cells/5 experiments), Control + BD1047 (92 cells/4 experiments). MCF7 cells TMRM measurements: Control (72 cells/4 experiments), Control + SKF10047 (111 cells/4 experiments) and Control + BD1047 (81 cells/4 experiments). A549 cells pyruvate to lactate ratio: Control (69 cells/6 experiments), Control + SKF10047 (60 cells/6 experiments), Control + BD1047 (69 cells/6 experiments), SR1 KD (58 cells/5 experiments), S1R KD + SKF10047 (49 cells/4 experiments) and S1RKD + BD1047 (45 cells/4 experiments). MCF7 cells pyruvate to lactate ratio: Control (54 cells/4 experiments), Control + SKF10047 (52 cells/4 experiments) and Control + BD1047 (53 cells/4 experiments). S1R-mCherry OE in MCF7 cells pyruvate to lactate ratio: Control (65 cells/8 experiments), Control + SKF10047 (83 cells/9 experiments) and Control + BD1047 (49 cells/7 experiments).
Next, we assessed the cytosolic pyruvate/lactate ratio with genetically encoded sensor Lapronic [11] (Figure 3D). Activation of S1R with the agonist resulted in increased pyruvate/lactate ratio in A549 cells and in MCF7 cells with S1R OE, but not in the case of S1R KD or MCF7 cells without OE (Figure 3 E–H). Interestingly, the antagonist, as well as S1R KD, reduced the pyruvate/lactate ratio in A549 cells (Figure 3E–G). Pyruvate/lactate ratio can be used to deduce the cell’s reliance on aerobic glycolysis versus Oxphos, as the increase of the ratio ((+)-SKF10047 in A549 and MCF7 with S1R OE) would suggest the cell is producing less lactate relative to pyruvate, and likely rerouting the pyruvate towards tricarboxylic acid (TCA) cycle. In case of decreased pyruvate/lactate ratio (BD1047 and S1R KD in A549 cells), the cell contains more lactate relative to pyruvate, suggesting it is performing more aerobic glycolysis. The pyruvate/lactate ratio measurements added a new perspective on the action of S1R under its basal, activated, and inactivated states. We have already shown that activation of S1R promotes OXPHOS. Based on pyruvate/lactate ratio, it also reduces aerobic glycolysis, while inactivation or KD of S1R seems to increase aerobic glycolysis, while not affecting mitochondrial ATP production or Ψm, suggesting basal involvement of S1R in balancing aerobic glycolysis and OXPHOS.
3.4. S1R Activation Boosts Mitochondrial Ca2+ Homeostasis
To investigate the mechanism of action of S1R activation and inactivation, we measured mitochondrial Ca2+ levels in A549 and MCF7 cells (Figure 4). (+)-SKF10047 treatment increased basal mitochondrial Ca2+ level as well as IP3generating agonist-induced mitochondrial Ca2+ uptake in control A549 cells (Figure 4A–C). This effect was abolished by S1R KD (Figure 4D–F) and was not present in MCF7 cells (Figure 4G–I). OE of S1R-mCherry construct in MCF7 cells mimicked the increased basal mitochondrial Ca2+ level induced by (+)-SKF10047, but not IP3 generating agonist-induced mitochondrial Ca2+ uptake, which was unaffected (Figure 4J–L). These results provide a mechanistic insight into Oxphos promoting effect of S1R activation, whereby (+)-SKF10047 promotes mitochondrial bioenergetics by increasing basal mitochondrial Ca2+ level as well as mitochondrial Ca2+ uptake, thus boosting both mitochondrial matrix residing Ca2+ sensitive NADH dehydrogenases along with mitochondrial intermembrane space (IMS) residing Ca2+ sensitive metabolite shuttles [17,18]. Together, these effects can theoretically overweight depolarizing effect of increased basal Ca2+ in mitochondria [20,21].
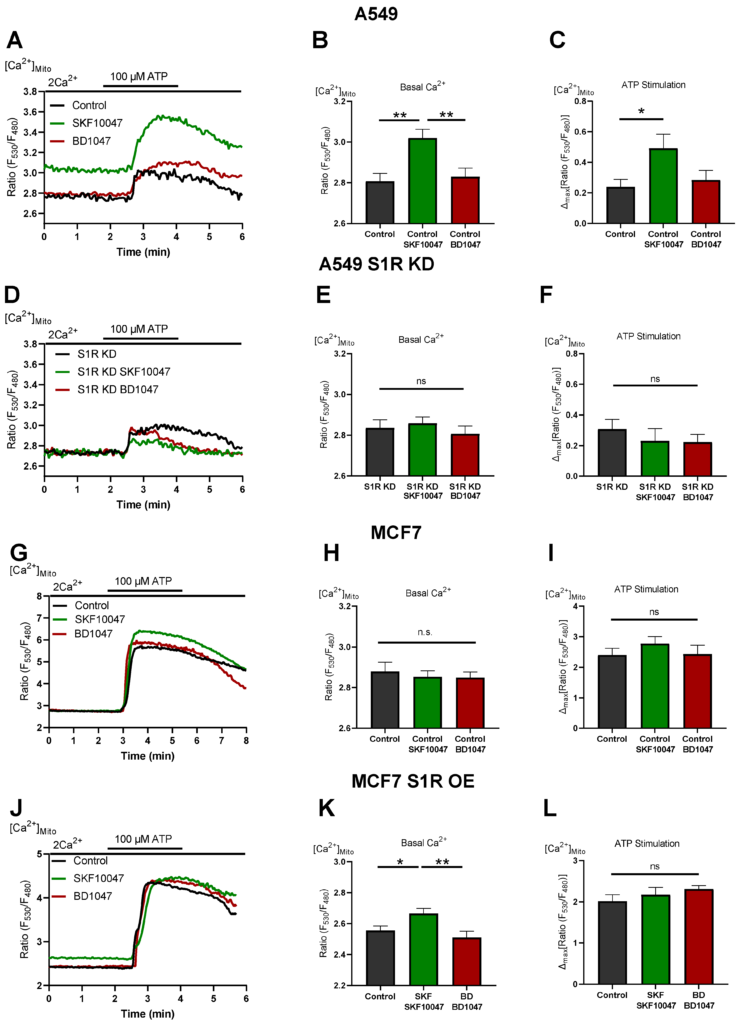
S1R activation increases basal and agonist-induced mitochondrial Ca2+ uptake.
(A) Representative traces of mitochondrial Ca2+ dynamics measured with 4mtD3cpv for control (black), Control+SKF10047 (green) and Control + BD1047 (red) in A549 cells. (B) Bar graphs with MEAN±SEM represent basal mitochondrial Ca2+ level for Control (black), Control+SKF10047 (green) and Control+BD1047 (red) in A549 cells. (C) Bar graphs with MEAN±SEM represent ATP (100 µM) induced mitochondrial Ca2+ uptake for Control (black), Control+SKF10047 (green) and Control+BD1047 (red) in A549 cells. (D) Representative traces of mitochondrial Ca2+ dynamics for S1R KD (black), S1R KD +SKF10047 (green) and S1R KD +BD1047 (red) in A549 cells. (E) Bar graphs with MEAN±SEM represent basal mitochondrial Ca2+ level for S1R KD (black), S1R KD +SKF10047 (green) and S1R KD +BD1047 (red) in A549 cells. (F) Bar graphs with MEAN±SEM represent ATP (100 µM) induced mitochondrial Ca2+ uptake for S1R KD (black), S1R KD +SKF10047 (green) and S1R KD +BD1047 (red) in A549 cells. (G) Representative traces of mitochondrial Ca2+ dynamics for Control (black), Control+SKF10047 (green) and Control+BD1047 (red) in MCF7 cells. (H) Bar graphs with MEAN±SEM represent basal mitochondrial Ca2+ level for Control (black), Control+SKF10047 (green) and Control+BD1047 (red) in MCF7 cells. (I) Bar graphs with MEAN±SEM represent ATP (100 µM) induced mitochondrial Ca2+ uptake for Control (black), Control+SKF10047 (green) and Control+BD1047 (red) in MCF7 cells. (J) Representative traces of mitochondrial Ca2+ dynamics for Control (black), Control+SKF10047 (green) and Control+BD1047 (red) in S1R-mCherry OE MCF7 cells. (K) Bar graphs with MEAN±SEM represent basal mitochondrial Ca2+ level for Control (black), Control+SKF10047 (green) and Control+BD1047 (red) in S1R-mCherry OE MCF7 cells. (L) Bar graphs with MEAN±SEM represent ATP (100 µM) induced mitochondrial Ca2+uptake for Control (black), Control+SKF10047 (green) and Control+BD1047 (red) in S1R-mCherry OE MCF7 cells. Cells were treated with BD1047 and SKF10047 2–4 h prior to each experiment. Significant differences were assessed using one-way ANOVA with Tukey’s multiple comparison test and presented as specific p-values (* p < 0.05, ** p < 0.01, ns: not significant). A549 cells basal Ca2+ measurements: Control (44 cells/9 experiments), Control + SKF10047 (44 cells/8 experiments), Control + BD1047 (38 cells/7 experiments), SR1 KD (32 cells/7 experiments), S1R KD + SKF10047 (43 cells/8 experiments) and S1RKD + BD1047 (25 cells/7 experiments). A549 cells ATP stimulation: Control (17 cells/9 experiments), Control + SKF10047 (20 cells/8 experiments), Control + BD1047 (18 cells/7 experiments), SR1 KD (11 cells/7 experiments), S1R KD + SKF10047 (12 cells/8 experiments) and S1RKD + BD1047 (9 cells/7 experiments). MCF7 cells basal Ca2+ measurements: Control (34 cells/3 experiments), Control + SKF10047 (54 cells/5 experiments) and Control + BD1047 (44 cells/4 experiments). MCF7 cells ATP stimulation: Control (16 cells/3 experiments), Control + SKF10047 (18 cells/5 experiments) and Control + BD1047 (20 cells/4 experiments). S1R-mCherry OE MCF7 cells basal Ca2+measurements: Control (26 cells/6 experiments), Control + SKF10047 (31 cells/6 experiments) and Control + BD1047 (22 cells/6 experiments). S1R-mCherry OE MCF7 cells ATP stimulation: Control (17 cells/6 experiments), Control + SKF10047 (17 cells/6 experiments) and Control + BD1047 (14 cells/6 experiments).
On the other hand, BD1047 had no effect on basal Ca2+ level as well as on mitochondrial Ca2+ uptake (Figure 4). Hence, mitochondrial Ca2+ data do not provide an explanation for increased aerobic glycolysis under S1R inactivation by BD1047 or S1R KD. None of the compounds had a considerable impact on basal or agonist-induced Ca2+ increase in the cytosol (Supplementary Figure S3).
4. Discussion
It is well known that cancer cells reprogram their metabolism to fit their demand for uncontrolled proliferation and survival [22]. Cancer energy metabolism is targeted by various anticancer treatments [23], but still requires a better understanding to design more efficient treatments strategies. In this study, we wanted to investigate the role of S1R in cancer energy metabolism. In particular, we were interested in basal, activated, and inactivated states of S1R. For this, we have chosen two cancer cell lines that differentially express S1R (Figure 1A,B) and treated them with established S1R agonist (+)-SKF10047 or antagonist BD1047. Additionally, we knocked-down S1R in cells expressing the protein (A549, Figure 1A–D) and overexpressed it in cells that have low levels of S1R (MCF7, Supplementary Figure S1), to test the specificity of the S1R ligands in their action on cancer energy metabolism.
Using real-time mitochondrial ATP, Ψm, and cytosolic pyruvate/lactate ratio measurements, we have established that activation of S1R by its agonist enhances OXPHOS and reduces reliance on aerobic glycolysis in A549 cells and in MCF7 cells transiently overexpressing S1R (Figure 2B,E and Figure 3B,E,H). In contrast, the S1R antagonist and S1R KD did not have an impact on OXPHOS, but increased aerobic glycolysis (Figure 2B,C and Figure 3B,E,F) in A549 cells, suggesting that S1R has a metabolic balancing function. Based on our results and previous publications [15], S1R seems to be rather dormant regarding its influence on mitochondrial bioenergetics under resting conditions, since S1R antagonist and S1R KD did not affect mitochondrial ATP and Ψm (Figure 2B,C and Figure 3E,F) and S1R OE in MCF7 cells did not drastically change metabolic phenotype of the cells unless treated with the agonist (Figure 2E and Figure 3H). As MCF7 serves as a control cell line with negligible S1R expression, none of the ligands impacted measured parameters in MCF7 cells (Figure 2D and Figure 3C,G).
We have recently shown that S1R is indispensable for enhancing mitochondrial bioenergetics during early ER stress by orchestrating ER Ca2+ leak towards mitochondria [15]. As it is known that ER stress activates S1R, we wanted to test if pharmacological activation of S1R also increases mitochondrial Ca2+ levels and thus explains boosted mitochondrial bioenergetics. Indeed, activation of S1R with (+)-SKF10047 increased both basal Ca2+ levels as well as mitochondrial Ca2+ uptake upon ER Ca2+ release in A549 cells (Figure 4A–C), while not affecting cytosolic Ca2+levels (Supplementary Figure S3A–C). This increase of basal Ca2+ level and increased ER Ca2+ release directed towards mitochondria are likely responsible for increased mitochondrial bioenergetics upon S1R activation, as Ca2+ is a known regulator of cellular and mitochondrial bioenergetics [20,24]. Interestingly, (+)-SKF10047 treatment of transiently S1R overexpressing MCF7 cells only increased basal mitochondrial Ca2+ and not mitochondrial Ca2+uptake upon ER Ca2+ release (Figure 4J–L). We speculate that this partial enhancement of mitochondrial Ca2+homeostasis is responsible for increased bioenergetics in agonist treated MCF7 cells with S1R OE and can be a reason the effect of the agonist is not as pronounced as in A549 cells, which endogenously express higher levels of S1R (Figure 1A,B and Figure 2).
Our findings are further supported by reports showing that ligand-activated S1R increases bradykinin-induced cytosolic Ca2+ rise in neuroblastoma cells by dissociating adaptor protein Ankyrin from IP3Rs [25]. Additionally, it was shown that activation of S1R via agonist stimulation leads to the interaction of S1R with IP3Rs and controls the prolonged Ca2+ delivery to the mitochondria [1].
It is still an open question whether S1R has a constitutively active function under basal conditions. We observed mixed results upon application of S1R antagonist or S1R KD. Both S1R antagonist and KD failed to impact mitochondrial bioenergetics (Figure 2B,C and Figure 3B), which is explainable in light of unaffected mitochondrial Ca2+ levels (Figure 4A–F). Although it was reported that S1R stabilizes IP3Rs, the effect of S1R KD was mainly reported to occur upon consecutive applications of IP3 generating agonist [1]. Thus, it is not surprising that we have not observed an effect of S1R KD on the initial Ca2+ release from the ER. On the other hand, decreased cytosolic pyruvate/lactate ratio in BD1047-treated or S1R KD cells (Figure 3E,F) argues for basal activity of S1R, which might be compensated by increased aerobic glycolysis upon S1R inactivation or KD. However, the mechanism of action in this case requires further clarification.